|
|
||
|
Since the CardiacSimu is maintained by GNU make, it is really simple to run cardiacsimu in a GNU Linux or Unix like system. Try $make all in the top of our source location. And all executable file of CardiacSimu is prepared in ./bin. But be caution, following libraries or tools should be installed before:
If CardiacSimu is built successfully in your computer, your bin directory should contains following files: executable binaries (hrteditor, slicer, solver, stl2geo, viewer), scripts (calcEcg.sh, dpl2isop.sh, dspisoc.sh, dspprop.sh, plotAP, plotEcg) and a configure file (project.config). If you want to use CardiacSimu in windows, you should use code::blocks + mingw to build slicer, solver and viewer. To build hrteditor, you should firstly download gtk+-2.x.x.x-bundle from the website and unpack it to C:\Program Files\gtk+-bundle. Then use code::blocks to build it. The code::blocks's project files are all in wincb folder. You could load cardiacsimu-win.workspace file into code::blocks to open them all. If CardiacSimu is built successfully, the executable binaries (hrteditor, slicer, solver, viewer) are in bin fold. The scripts and configure file should be manually copied into bin fold. And in windows, gnuplot script could not be used. Fortunately, there are dozens of tools could be used to explore data in csv file. You could use msys to invoke corresponding tool (solver etc.) to do simulation. | ||
|
|
||
|
To run simulation, you should have a anatomical model represented as stl file in ./bin, and right now you can find one here. |
||
|
|
||
|
configure file is in your ./bin fold named project.config, if not, just copy it from ./config to ./bin. See the figure in the left, the grey one is the original anatomical model and red one, composed by many little sphere elements, is the partition result. |
||
|
|
||
|
Right figure indicates the definition used in my research, according to some literature (Harrild 2000), we defined SN (sinus node, little grey part connect BB and CT), BB (bachman bundle, blue fiber), FOL (fossa ovalis limbus, green fiber in circle), FO (fossa ovalis, red parts in FOL), Pects (pectinate muscle, purple), CT(crista terminalis, yellow fiber connected all pects), IB (intercaval bundle, yellow fiber connect FOL and CT) and Isthmus (right atria floor isthmus, red parts at the bottom of right atrium). |
||
|
|
||
|
You can find these configurable items in simulation segment in configure file :). |
||
|
|
||
|
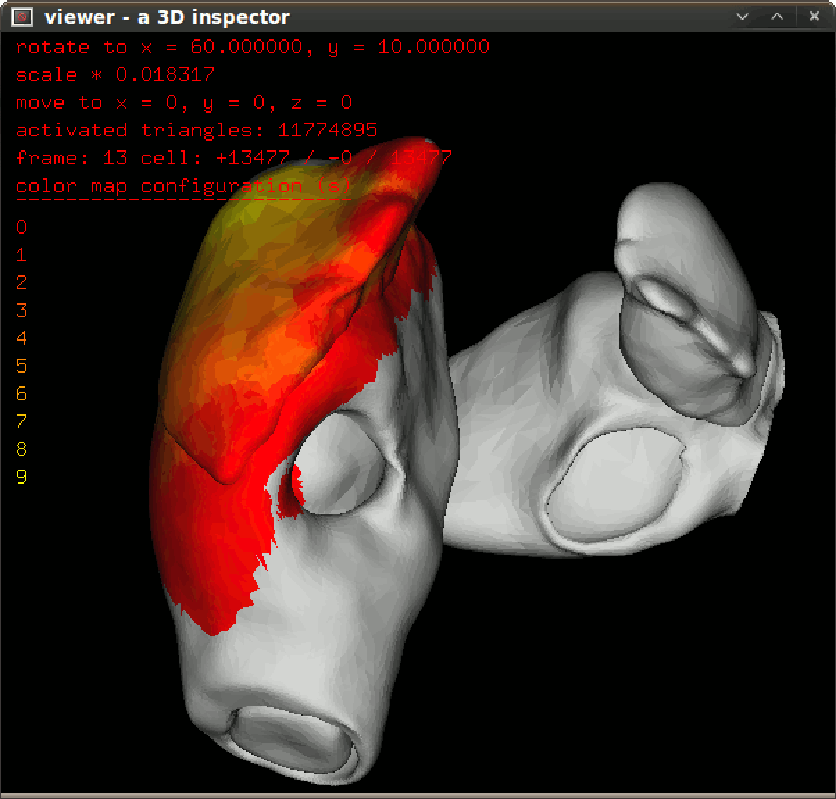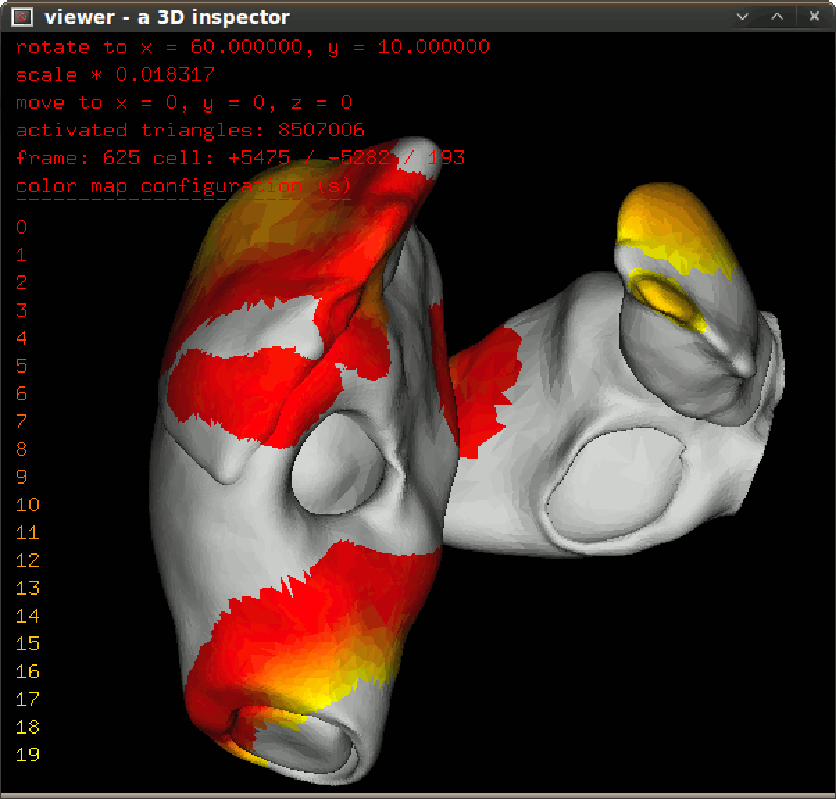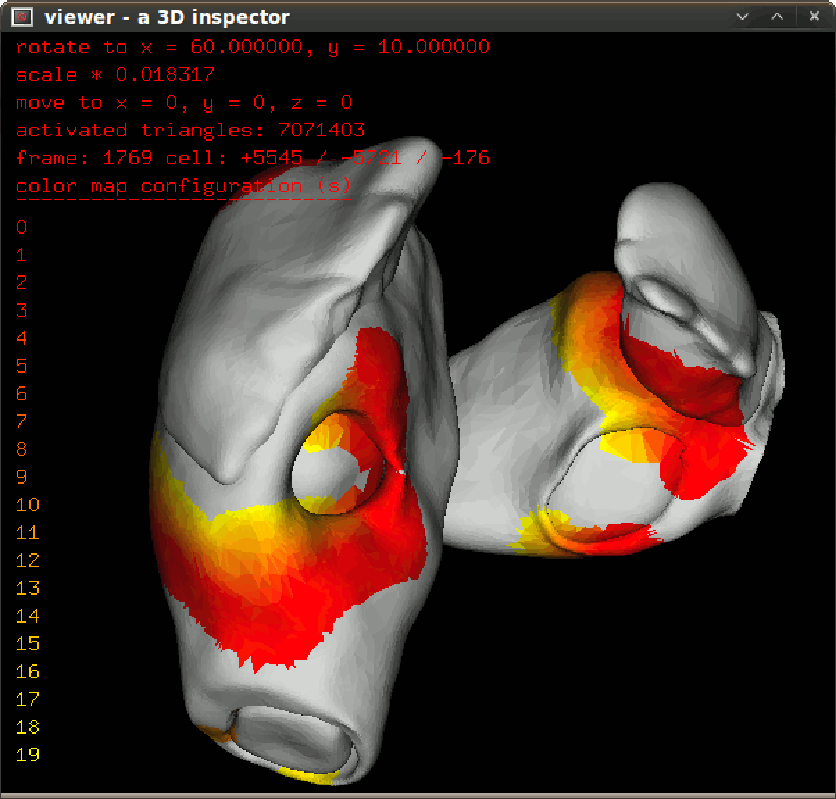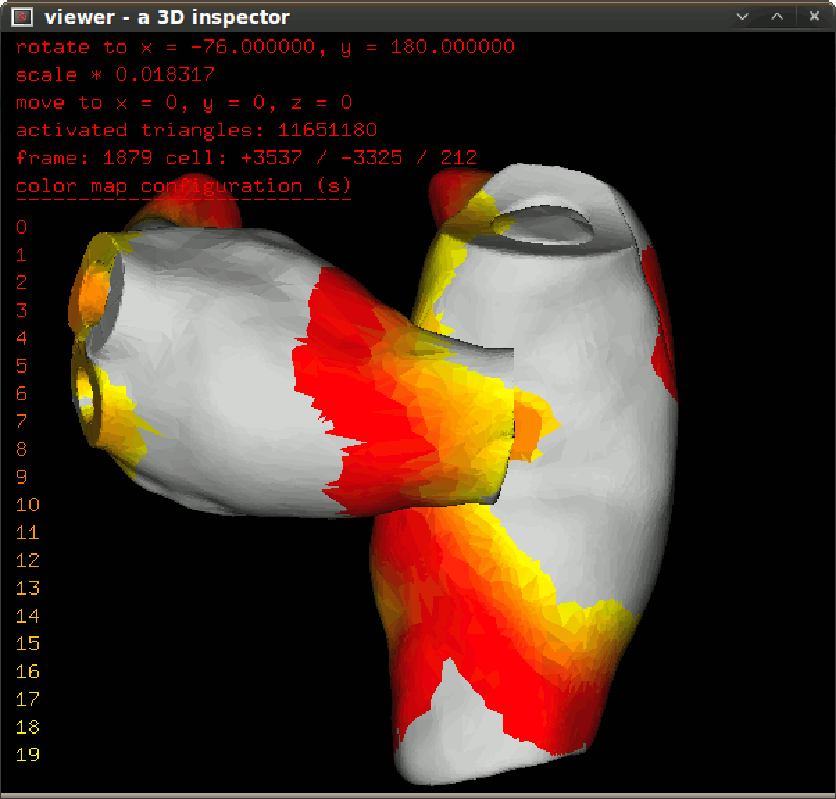
To inspect activation propagation in a transparent 3D scene use $bash dspprop.sh. The figure in left give the propagation snapshot in 10ms step. To inspect isochronal map in a transparent 3D scene, use $bash dspisoc.sh. The figure in right give the isochronic maps in a reentry condition. |
||
|
|
||
|
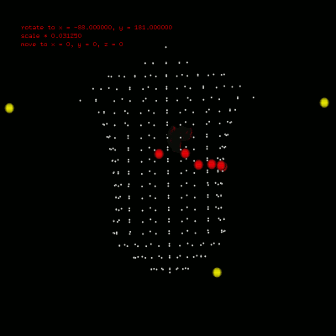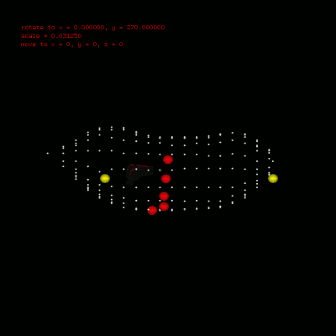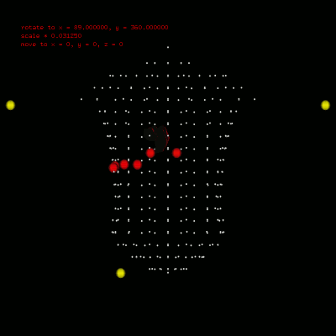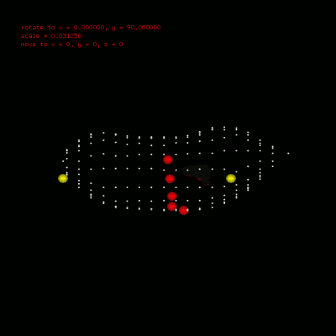
To calculate ecg, you should also define the leads position, which can also configured in anatomical segment (anatomical::limbtriindex and anatomical::precordialtriindex). anatomical::limbtriindex defines the xyz coordination of limb leads (left arm in la, right arm in ra and left leg in ll). anatomical::precordialtriindex defines the triangle mesh index of torso model for each precordial lead to settle. To view the location result you need used $viewer -stl stlfile -torso. Left figure gives the location configuration in my research. Silver spheres are the vertex of torso model, red spheres are the precordial leads and yellow ones are the limb leads. Till now, you can use $bash calcEcg.sh to calculate ECG waveform on limb leads and precordial leads. The ECG waveform is stored in csv file, you can use any software (Excel, Matlab etc) to investigate. Or use $gnuplot plotEcg to export ECG waveform in ecg.png. |
||
|
|
||
|
CardiacSimu will only existed in the directory where you initial keep the download archive, and whole software will be remove if you delete this directory. | ||
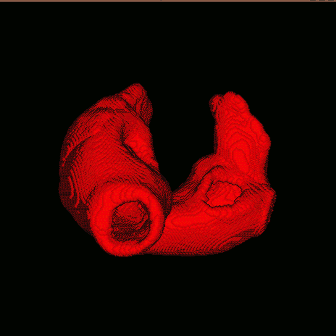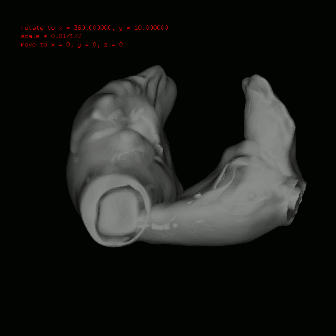 use $slicer stlfile to get simulating model (in hrt file shared the same name with stl file), before the partition, you can define the scale on i,j,k coordination axis in element segment (element::scale) in configure file. After the partition, you should copy the radius of sphere element into element segment (element::radius).
use $slicer stlfile to get simulating model (in hrt file shared the same name with stl file), before the partition, you can define the scale on i,j,k coordination axis in element segment (element::scale) in configure file. After the partition, you should copy the radius of sphere element into element segment (element::radius).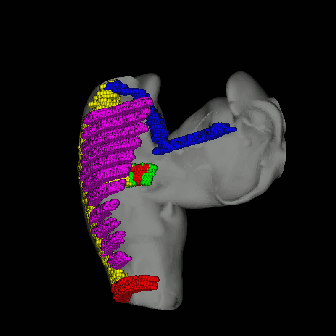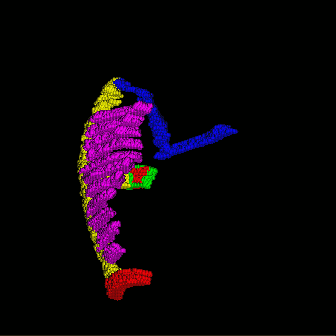 use $hrteditor hrtfile to define special conduction system in simulating model and use $viewer -hrt hrtfile and $viewer -stl stlfile -hrt hrtfile to investigate results. You can investigate the location of special conducting system by using viewer with -hrt argument.
use $hrteditor hrtfile to define special conduction system in simulating model and use $viewer -hrt hrtfile and $viewer -stl stlfile -hrt hrtfile to investigate results. You can investigate the location of special conducting system by using viewer with -hrt argument.
 use $solver -propagation-simu to simulate propagation. When this step is fulfilled, you can find several dpl file and xct file in ./bin directory. Be caution, you should leave enough disk free space for the propagation simulation.
use $solver -propagation-simu to simulate propagation. When this step is fulfilled, you can find several dpl file and xct file in ./bin directory. Be caution, you should leave enough disk free space for the propagation simulation.